My name is Rich. I live in New Jersey. My hobbies include genealogy and ruining home improvement projects. My favorite color is green, but it used to be blue. My favorite book is Catch-22, and my favorite sport is spreadsheets (so... baseball).


I'm currently a staff researcher in the Blekhman Lab at the University of Chicago, where we study the human microbiome. My work is focused on gut microbes and how they interact with their hosts, and how we can use big data and machine learning to extract more meaning from noisy ecological data. Previously, I was a bioinformatician in the labs of Drs. Rajan Jain and Eric Joyce, where I worked on epigenetics and nuclear architecture.
I got my PhD from the University of Minnesota. Before that, I worked as a software developer at the Minnesota Supercomputing Institute, and at companies including USA Today, Target and Amazon Web Services.
All my publications are on my Google Scholar page, but the papers that best describe my research interests are here.
My primary field of study is the human gut microbiome, especially novel approaches to the analysis of sequencing-based assays. I'm a leader of the Human Microbiome Compendium project.
Abdill RJ*, Graham SP*, Rubinetti V, Ahmadian M, Hicks P, Chetty A, McDonald D, Ferretti P, Gibbons E, Rossi M, Krishan A, Albert FW, Greene CS, Davis S, Blekhman R.
Cell (2025)

The factors shaping human microbiome variation are a major focus of biomedical research. While other fields have used large sequencing compendia to extract insights requiring otherwise impractical sample sizes, the microbiome field has lacked a comparably sized resource for the 16S rRNA gene amplicon sequencing commonly used to quantify microbiome composition. To address this gap, we processed 168,464 publicly available human gut microbiome samples with a uniform pipeline. We use this compendium to evaluate geographic and technical effects on microbiome variation. We find that regions such as Central and Southern Asia differ significantly from the more thoroughly characterized microbiomes of Europe and Northern America and that composition alone can be used to predict a sample’s region of origin. We also find strong associations between microbiome variation and technical factors such as primers and DNA extraction. We anticipate this growing work, the Human Microbiome Compendium, will enable advanced applied and methodological research.
Abdill RJ, Adamowicz EM & Blekhman R
PLOS Biology (2022)
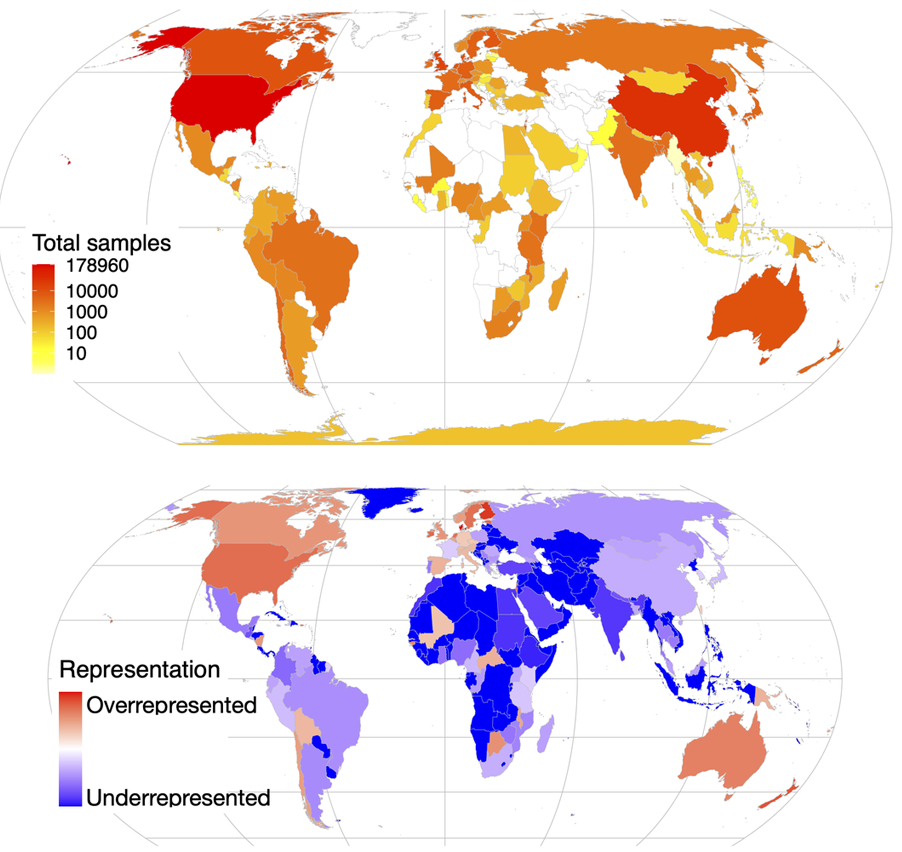
The importance of sampling from globally representative populations has been well established in human genomics. In human microbiome research, however, we lack a full understanding of the global distribution of sampling in research studies. This information is crucial to better understand global patterns of microbiome-associated diseases and to extend the health benefits of this research to all populations. Here, we analyze the country of origin of all 444,829 human microbiome samples that have been collected to date and are available from the world’s three largest genomic data repositories, including the Sequence Read Archive (SRA). We show that more than 71% of publicly available human microbiome samples with a known origin come from Europe, the United States, and Canada, including 46.8% from the United States alone, despite the country representing only 4.3% of the global population. We also find that central and southern Asia is the most underrepresented region: Countries such as India, Pakistan, and Bangladesh account for more than a quarter of the world population but make up only 1.8 percent of human microbiome samples. These results demonstrate a critical need to ensure more global representation of participants in microbiome studies.
I'm very interested in the "science of science" and have done work collecting and evaluating metadata from the bioRxiv and medRxiv preprint servers, PubMed Central, and NCBI databases.
Abdill RJ, Adamowicz EM & Blekhman R
eLife (2020)
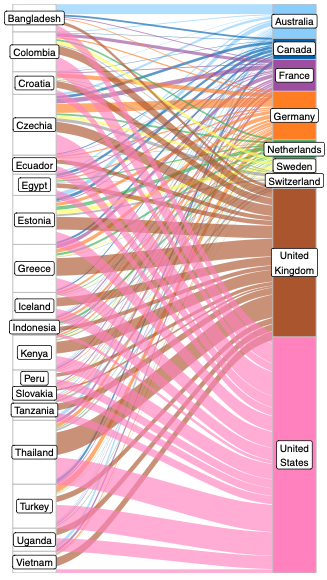
Preprints are becoming well established in the life sciences, but relatively little is known about the demographics of the researchers who post preprints and those who do not, or about the collaborations between preprint authors. Here, based on an analysis of 67,885 preprints posted on bioRxiv, we find that some countries, notably the United States and the United Kingdom, are overrepresented on bioRxiv relative to their overall scientific output, while other countries (including China, Russia, and Turkey) show lower levels of bioRxiv adoption. We also describe a set of 'contributor countries' (including Uganda, Croatia and Thailand): researchers from these countries appear almost exclusively as non-senior authors on international collaborations. Lastly, we find multiple journals that publish a disproportionate number of preprints from some countries, a dynamic that almost always benefits manuscripts from the US.
Abdill RJ & Blekhman R
PLOS Biology (2019)
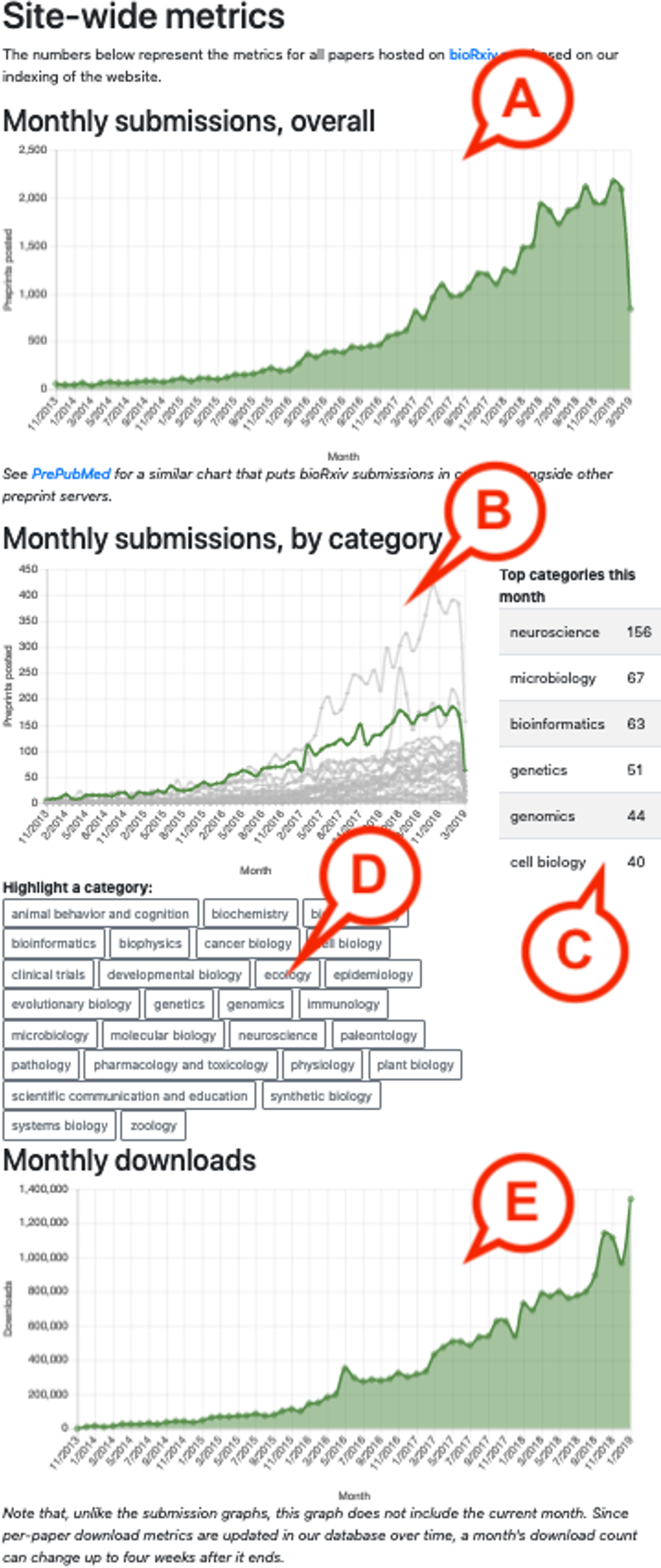
Preprints have arrived. In increasing numbers, researchers across the life sciences are embracing the once-niche practice, shaking off decades of reluctance and posting hundreds of papers per week to preprint servers, sharing their findings with the community before embarking on the weary march through peer review. However, there are limited methods for individuals sifting through this avalanche of research to identify the preprints that are most relevant to their interests. Here, we describe Rxivist.org, a website that indexes all preprints posted to bioRxiv.org, the largest preprint server in the life sciences, and allows users to filter and sort papers based on download metrics and Twitter activity over a variety of categories and time periods. In this work, we hope to make it easier for readers to find relevant research on bioRxiv and to improve the visibility of preprints currently being read and discussed online.
Abdill RJ & Blekhman R
eLife (2019)
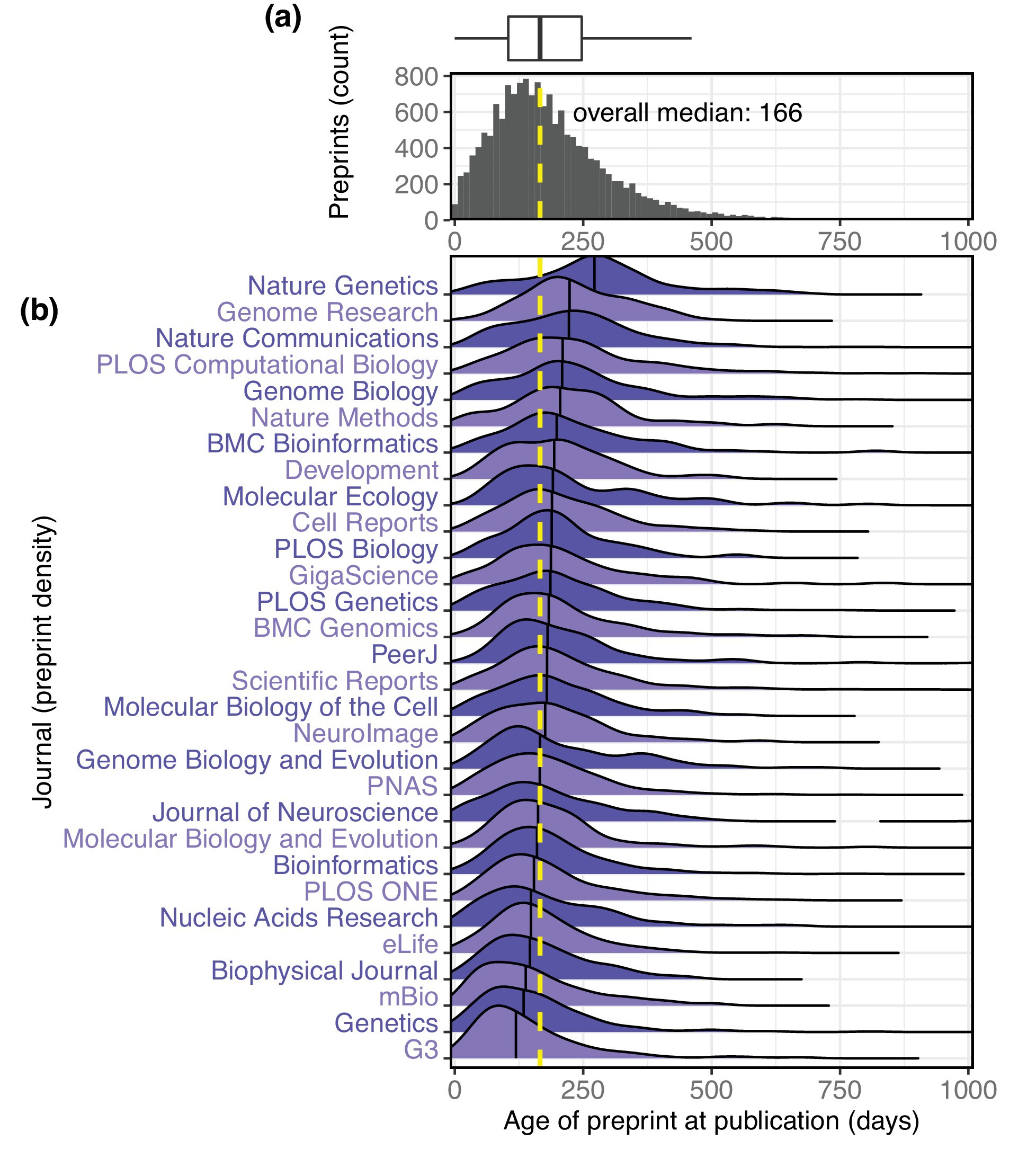
The growth of preprints in the life sciences has been reported widely and is driving policy changes for journals and funders, but little quantitative information has been published about preprint usage. Here, we report how we collected and analyzed data on all 37,648 preprints uploaded to bioRxiv.org, the largest biology-focused preprint server, in its first five years. The rate of preprint uploads to bioRxiv continues to grow (exceeding 2,100 in October 2018), as does the number of downloads (1.1 million in October 2018). We also find that two-thirds of preprints posted before 2017 were later published in peer-reviewed journals, and find a relationship between the number of downloads a preprint has received and the impact factor of the journal in which it is published. We also describe Rxivist.org, a web application that provides multiple ways to interact with preprint metadata.